Most Comprehensive Reference List Available Shows Impressive Depth, Breadth
Since about 2005 — and at an accelerating pace — Wikipedia has emerged as the leading online knowledge base for conducting semantic Web and related research. The system is being tapped for both data and structure. Wikipedia has arguably replaced WordNet as the leading lexicon for concepts and relations. Because of its scope and popularity, many argue that Wikipedia is emerging as the de facto structure for classifying and organizing knowledge in the 21st century.
Our work on the UMBEL lightweight reference subject concept structure has stated since the project’s announcement in July 2007 that Wikipedia is a key intended resource for identifying subject concepts and entities. For the past few months I have been scouring the globe attempting to find every drop of research I could find on the use of Wikipedia for semantic Web, information extraction, categorization and related issues.
Thus, I’m pleased to offer up herein the most comprehensive such listing available anywhere: more than 99 resources and counting! (I say “more than” because some entries below have multiple resources; I just liked the sound of 99 as a round number!)
Wikipedia itself maintains a listing of academic studies using Wikipedia as a resource; fewer than one-third of the listings below are on that list (which itself may be an indication of the current state of completeness within Wikipedia). Some bloggers and other sources around the Web also maintain listings in lesser degrees of completeness.
It is well documented the tremendous growth of content and topics within Wikipedia (see, as examples, the W1, W2, W3, W4, W5, W6 and W7 internal Wikipedia sources for gory details), with as of early 2008 about 2.25 million articles in English and versions in 256 languages and variants.
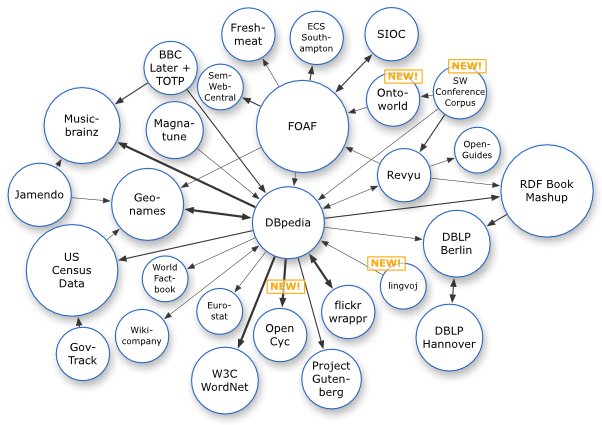
Download access to the full knowledge base has enabled the development of notable core references to the Linked Data aspects of the semantic Web such as DBpedia [5,6] and YAGO [72,73]. Entire research teams, such as Ponzetto and Strube [61-65] (and others as well; see below) are moving toward creating a full-blown ontologies or structured knowledge bases useful for semantic Web purposes based on Wikipedia. So, one of the first and principle uses of Wikipedia to date has been as a data source of concepts, entities and relations.
But much broader data mining and text mining and analysis is being conducted against Wikipedia, that is currently defining the state-of-the-art in these areas, too:
- Ontology development and categorization
- Word sense disambiguation
- Named entity recognition
- Named entity disambiguation
- Semantic relatedness and relations.
These objectives, in turn, are mining and extracting these various kinds of structure for these purposes in Wikipedia:
- Articles
- First paragraph — Definitions
- Full text — Description of meaning; related terms; translations
- Redirects — Synonymy; spelling variations, misspellings; abbreviations
- Title — Named entities; domain specific terms or senses
- Subject — Category suggestion (phrase marked in bold or in first paragraph)
- Section heading — Category suggestions
- Article links
- Context — Related terms; co-occurrences
- Label — Synonyms; spelling variations; related terms
- Target — Link graph; related terms
- LinksTo — Category suggestion
- LinkedBy — Category suggestion
- Categories
- Category — Category suggestion
- Contained articles — Semantically related terms (siblings)
- Hierarchy — Hyponymic and meronymic relations between terms
- Disambiguation pages
- Article links — Sense inventory
- Infobox Templates
- Name —
- Item — Category suggestion; entity suggestion
- Lists
- Hyponyms
These are some of the specific uses that are included in the 99 resources listed below.
This is an exciting (and, for most all of us just a few years back, unanticipated) use of the Web in socially relevant and contextual knowledge and research. I’m sure such a listing one year by now will be double in size or larger!
BTW, suggestions for new or overlooked entries are very much welcomed! 🙂
A – B
- Sisay Fissaha Adafre and Maarten de Rijke, 2006. Finding Similar Sentences across Multiple Languages in Wikipedia, in EACL 2006 Workshop on New Text–Wikis and Blogs and Other Dynamic Text Sources, April 2006.See http://www.science.uva.nl/~mdr/Publications/Files/eacl2006-similarsentences.pdf.
- Sisay Fissaha Adafre, V. Jijkoun and M. de Rijke. Fact Discovery in Wikipedia, in 2007 IEEE/WIC/ACM International Conference on Web Intelligence. See http://staff.science.uva.nl/~mdr/Publications/Files/wi2007.pdf.
- Sisay Fissaha Adafre and Maarten de Rijke, 2005. Discovering Missing Links in Wikipedia, in LinkKDD 2005, August 21, 2005, Chicago, IL. See http://data.isi.edu/conferences/linkkdd-05/Download/Papers/linkkdd05-13.pdf.
- David Ahn, Valentin Jijkoun, Gilad Mishne, Karin Müller, Maarten de Rijke, and Stefan Schlobach. 2004. Using Wikipedia at the TREC QA Track, in Proceedings of TREC 2004.
- Sören Auer, Chris Bizer, Jens Lehmann, Georgi Kobilarov, Richard Cyganiak and Zachary Ives, 2007. DBpedia: A nucleus for a web of open data, in Proceedings of the 6th International Semantic Web Conference and 2nd Asian Semantic Web Conference (ISWC/ASWC2007), Busan, South Korea, volume 4825 of LNCS, pages 715–728, November 2007. See http://iswc2007.semanticweb.org/papers/ISWC2007_IU_Auer.pdf.
- Sören Auer and Jens Lehmann, 2007. What Have Innsbruck and Leipzig in Common? Extracting Semantics from Wiki Content, in The Semantic Web: Research and Applications, pages 503-517, 2007. See http://www.eswc2007.org/pdf/eswc07-auer.pdf.
- Somnath Banerjee, 2007. Boosting Inductive Transfer for Text Classification Using Wikipedia, at the Sixth International Conference on Machine Learning and Applications (ICMLA). See http://portal.acm.org/citation.cfm?id=1336953.1337115.
- Somnath Banerjee, Krishnan Ramanathan, Ajay Gupta, 2007. Clustering Short Texts using Wikipedia, poster presented at Proceedings of the 30th Annual International ACM SIGIR Conference on Research and Development in Information Retrieval, Amsterdam, The Netherlands, pp. 787-788.
- F. Bellomi and R. Bonato, 2005. Lexical Authorities in an Encyclopedic Corpus: A Case Study with Wikipedia, online reference not found.
- F. Bellomi and R. Bonato, 2005. Network Analysis for Wikipedia, presented at Wikimania 2005; see http://www.fran.it/articles/wikimania_bellomi_bonato.pdf
- Bibauw (2005) analyzed the lexicographical structure of Wiktionary (in French).
- Abhijit Bhole, Blaž Fortuna, Marko Grobelnik and Dunja Mladenić, 2007.Mining Wikipedia and Relating Named Entities over Time, see http://www.cse.iitb.ac.in/~abhijit.bhole/SiKDD2007ExtractingWikipedia.pdf.
- Razvan Bunescu and Marius Pasca. 2006. Using Encyclopedic Knowledge for Named Entity Disambiguation, in Proceedings of the 11th Conference of the EACL, pages 9-16, Trento, Italy.
- Razvan Bunescu, 2007. Learning for Information Extraction: From Named Entity Recognition and Disambiguation To Relation Extraction, Ph.D. thesis for the University of Texas, August 2007, 168 pp. See http://oucsace.cs.ohiou.edu/~razvan/papers/thesis-white.pdf.
- Luciana Buriol, Carlos Castillo, Debora Donato, Stefano Leonardi, and Stefano Millozzi. 2006. Temporal Analysis of the Wikigraph, in Proceedings of Web Intelligence, Hong Kong.
- Davide Buscaldi and Paolo Rosso, 2007. A Comparison of Methods for the Automatic Identification of Locations in Wikipedia, in Proceedings of GIR’07, November 9, 2007, Lisbon, Portugal, pp 89-91. See http://www.dsic.upv.es/~prosso/resources/BuscaldiRosso_GIR07.pdf.
C – F
- Ruiz-Casado, M., Alfonseca, E., and Castells, P. 2005. Automatic Assignment of Wikipedia Encyclopedic Entries to WordNet Synsets, in AWIC, pages 380-386. See http://nets.ii.uam.es/publications/nlp/awic05.pdf.
- Maria Ruiz-Casado, Enrique Alfonseca and Pablo Castells, 2006. From Wikipedia to Semantic Relationships: a Semi-automated Annotation Approach, in ESWC2006.
- Maria Ruiz-Casado, Enrique Alfonseca and Pablo Castells, 2007. Automatising the Learning of Lexical Patterns: an Application to the Enrichment of WordNet by Extracting Semantic Relationships from Wikipedia. See http://nets.ii.uam.es/publications/nlp/dke07.pdf.
- Sergey Chernov, Tereza Iofciu, Wolfgang Nejdl, and Xuan Zhou, 2006. Extracting Semantic Relationships between Wikipedia Categories, from SEMWIKI 2006.
- Aron Culotta, Andrew McCallum and Jonathan Betz, 2006. Integrating Probabilistic Extraction Models and Data Mining to Discover Relations and Patterns in Text, in Proceedings of HLT-NAACL-2006. See http://www.cs.umass.edu/~culotta/pubs/culotta06integrating.pdf
- Silviu Cucerzan, 2007. Large-Scale Named Entity Disambiguation Based on Wikipedia Data, in Proceedings of the 2007 Joint Conference on Empirical Methods in Natural Language Processing and Computational Natural Language Learning (EMNLP-CoNLL). See http://www.aclweb.org/anthology-new/D/D07/D07-1074.pdf.
- Cyclopedia, from the Cyc Foundation, an online version of Wikipedia that enables browsing the encyclopedia by concepts. See http://www.cycfoundation.org/blog/?page_id=15.
- Wisam Dakka and Silviu Cucerzan, 2008. Augmenting Wikipedia with Named Entity Tags, to be published in IJCNLP. See http://research.microsoft.com/users/silviu/Papers/np-ijcnlp08.pdf.
- Wisam Dakka and Silviu Cucerzan, 2008. Also, see the online tool http://wikinet.stern.nyu.edu/ (not yet implemented).
- Turdakov Denis, 2007. Recommender System Based on User-generated Content. See http://syrcodis.citforum.ru/2007/9.pdf.
- EachWiki, online system from Fu et al.
- Linyun Fu, Haofen Wang, Haiping Zhu, Huajie Zhang, Yang Wang and Yong Yu, 2007. Making More Wikipedians: Facilitating Semantics Reuse for Wikipedia Authoring, at ISWC 2007. See http://iswc2007.semanticweb.org/papers/ISWC2007_RT_Fu.pdf.
G – H
- Evgeniy Gabrilovich and Shaul Markovitch. 2006. Overcoming the Brittleness Bottleneck using Wikipedia: Enhancing Text Categorization with Encyclopedic Knowledge, in AAAI, pages 1301-1306, Boston, MA.
- Evgeniy Gabrilovich and Shaul Markovitch. 2007. Computing Semantic Relatedness using Wikipedia-based Explicit Semantic Analysis, in Proceedings of the 20th International Joint Conference on Artificial Intelligence (IJCAI), Hyderabad, India, January 2007.
- Evgeniy Gabrilovich, 2006. Feature Generation for Textual Information Using World Knowledge, Ph.D. Thesis for The Technion – Israel Institute of Technology, Haifa, Israel, December 2006, 218 pp. See http://www.cs.technion.ac.il/~gabr/papers/phd-thesis.pdf. (There is also an informative video at http://www.researchchannel.org/prog/displayevent.aspx?rID=4915).)
- See also Gabrilovich’s Perl Wikipedia tool, WikiPrep.
- Rudiger Gleim, Alexander Mehler and Matthias Dehmer, 2007. Web Corpus Mining by Instance of Wikipedia, in Proc. 2nd Web as Corpus Workshop at EACL 2006. See http://acl.ldc.upenn.edu/W/W06/W06-1710.pdf.
- Andrew Gregorowicz and Mark A. Kramer, 2006. Mining a Large-Scale Term-Concept Network from Wikipedia, Mitre Technical Report, October 2006. See http://www.mitre.org/work/tech_papers/tech_papers_06/06_1028/06_1028.pdf.
- Andreas Harth, Hannes Gassert, Ina O’Murchu, John Breslin and Stefan Decker, 2005. WikiOnt: An Ontology for Describing and Exchanging Wiki Articles, presented at Wikimania, Frankfurt, 5th August 2005. See http://sw.deri.org/~jbreslin/presentations/20050805a.pdf.
- Martin Hepp, Daniel Bachlechner and Katharina Siorpaes, 2006. Harvesting Wiki Consensus – Using Wikipedia Entries as Ontology Elements, See http://www.heppnetz.de/files/SemWiki2006-Harvesting%20Wiki%20Consensus-LNCS-final.pdf.
- Martin Hepp, Katharina Siorpaes, Daniel Bachlechner, 2007. Harvesting Wiki Consensus: Using Wikipedia Entries as Vocabulary for Knowledge Management, in IEEE Internet Computing, Vol. 11, No. 5, pp. 54-65, Sept-Oct 2007. See http://www.heppnetz.de/files/hepp-siorpaes-bachlechner-harvesting%20wikipedia%20w5054.pdf. Also, sample data at http://www.heppnetz.de/harvesting-wikipedia/.
- A. Herbelot and Ann Copestake, 2006. Acquiring Ontological Relationships from Wikipedia Using RMRS, in Proc. International Semantic Web Conference 2006 Workshop, Web Content Mining with Human Language Technologies, Athens, GA, 2006. See http://orestes.ii.uam.es/workshop/12.pdf.
- Ryuichiro Higashinaka, Kohji Dohsaka and Hideki Isozaki, 2007. Learning to Rank Definitions to Generate Quizzes for Interactive Information Presentation, in Companion Volume to the Proceedings of the 45th Annual Meeting of the Association for Computational Linguistics; see pages 117-120 within http://acl.ldc.upenn.edu/P/P07/P07-2.pdf
- Todd Holloway, Miran Bozicevic, and Katy Börner. 2005. Analyzing and Visualizing the Semantic Coverage of Wikipedia and Its Authors. ArXiv Computer Science e-prints, cs/0512085.
- Wei Che Huang, Andrew Trotman, and Shlomo Geva, 2007. Collaborative Knowledge Management: Evaluation of Automated Link Discovery in the Wikipedia, in SIGIR 2007 Workshop on Focused Retrieval, July 27, 2007, Amsterdam, The Netherlands. See http://www.cs.otago.ac.nz/sigirfocus/paper_15.pdf.
I – K
- Jonathan Isbell and Mark H. Butler, 2007. Extracting and Re-using Structured Data from Wikis, Hewlett-Packard Technical Report HPL-2007-182, 14th November, 2007, 22 pp. See http://www.hpl.hp.com/techreports/2007/HPL-2007-182.pdf.
- Maciej Janik and Krys Kochut, 2007. Wikipedia in Action: Ontological Knowledge in Text Categorization, University of Georgia, Computer Science Department Technical Report no. UGA-CS-TR-07-001. See http://lsdis.cs.uga.edu/~mjanik/UGA-CS-TR-07-001.pdf.
- Gjergji Kasneci, Fabian M. Suchanek, Georgiana Ifrim, Maya Ramanath and Gerhard Weikum, 2007. NAGA: Searching and Ranking Knowledge, Technical Report, Max-Planck-Institut f¨ur Informatik, MPI–I–2007–5–001, March 2007, 42 pp.See http://www.mpi-inf.mpg.de/~kasneci/naga/report.pdf
- Jun’ichi Kazama and Kentaro Torisawa, 2007. Exploiting Wikipedia as External Knowledge for Named Entity Recognition, in Proceedings of the 2007 Joint Conference on Empirical Methods in Natural Language Processing and Computational Natural Language Learning, pp. 698–707, Prague, June 2007.See http://acl.ldc.upenn.edu/D/D07/D07-1073.pdf.
- Daniel Kinzler, 2005. WikiSense Mining the Wiki, V 1.1, presented at Wikimania 2005, 10 pp. See http://brightbyte.de/repos/papers/2005/WikiSense-Presentation.pdf.
- A. Krizhanovsky, 2006. Synonym Search in Wikipedia: Synarcher, in 11th International Conference “Speech and Computer” SPECOM’2006. Russia, St. Petersburg, June 25-29, 2006, pp. 474-477. See http://arxiv.org/abs/cs/0606097 (also PDF).
- Natalia Kozlova, 2005. Automatic Ontology Extraction for Document Classification, a Master’s Thesis for Saarland University, February 2005, 90 pp. See http://domino.mpi-inf.mpg.de/imprs/imprspubl.nsf/80255f02006a559a80255ef20056fc02/3b864d86612739b0c1256fb70042547a/$FILE/Masterarbeit-Kozlova-Nat-2005.pdf. [READ]
- M. Krötzsch, D. Vrandečić and M. Völkel, 2005. Wikipedia and the Semantic Web: the Missing Links, at Wikimania 2005.Wikipedia and the Semantic Web: the Missing Links, at Wikimania 2005. See http://citeseer.ist.psu.edu/cache/papers/cs2/143/http:zSzzSzwww.aifb.uni-karlsruhe.dezSzWBSzSzmakzSzpubzSzwikimania.pdf/krotzsch05wikipedia.pdf.
L – N
- Rada Mihalcea and Andras Csomai, 2007. Wikify! Linking Documents to Encyclopedic Knowledge, in Proceedings of the Sixteenth ACM conference on Conference on information and knowledge management CIKM ’07 , November 6-8, 2007, pp. 233-241. See ACM portal retrieval (http://portal.acm.org/citation.cfm?id=1321475&coll=Portal&dl=ACM&CFID=8333672&CFTOKEN=48251251).
- Rada Mihalcea, 2007. Using Wikipedia for Automatic Word Sense Disambiguation, in Proceedings of NAACL HLT 2007, pages 196–203, April 2007.See http://www.cs.unt.edu/~rada/papers/mihalcea.naacl07.pdf.
- David Milne, 2007. Computing Semantic Relatedness using Wikipedia Link Structure; see also the Wikipedia Miner Toolkit (http://sourceforge.net/projects/wikipedia-miner/) provided by the author
- D. Milne, O. Medelyan and I. H. Witten, 2006. Mining Domain-Specific Thesauri from Wikipedia: A Case Study, in Proceedings of the International Conference on Web Intelligence (IEEE/WIC/ACM WI’2006), Hong Kong. Also Olena Medelyan’s home page, http://www.cs.waikato.ac.nz/~olena/.
- David Milne, Ian H. Witten and David M. Nichols, 2007. A Knowledge-Based Search Engine Powered by Wikipedia, at CIKM ’07.
- Lev Muchnik, Royi Itzhack, Sorin Solomon and Yoram Louzoun, 2007. Self-emergence of Knowledge Trees: Extraction of the Wikipedia Hierarchies, in Physical Review E (Statistical, Nonlinear, and Soft Matter Physics), Vol. 76, No. 1.
- Nadeau, D., Turney, P., Matwin, S., 2006. Unsupervised Named-Entity Recognition: Generating Gazetteers and Resolving Ambiguity, at 19th Canadian Conference on Artificial Intelligence. Québec City, Québec, Canada. June 7, 2006.See http://www.iit-iti.nrc-cnrc.gc.ca/iit-publications-iti/docs/NRC-48727.pdf. Doesn’t specifically use Wikipedia, but techniques are applicable.
- Kotaro Nakayama, Takahiro Hara and Shojiro Nishio, 2007. Wikipedia Mining for an Association Web Thesaurus Construction, in Web Information Systems Engineering – WISE 2007, Vol. 4831 (2007), pp. 322-334.
- Dat P.T. Nguyen, Yutaka Matsuo and Mitsuru Ishizuka, 2007. Relation Extraction from Wikipedia Using Subtree Mining, from AAAI ’07.
O – P
- Yann Ollivier and Pierre Senellar, 2007. Finding Related Pages Using Green Measures: An Illustration with Wikipedia, in Proceedings of the AAAI-07 Conference. See http://pierre.senellart.com/publications/ollivier2006finding.pdf. See also http://pierre.senellart.com/publications/ollivier2006finding/ for tools and data.
- Simon Overell and Stefan Ruger, 2006. Identifying and Grounding Descriptions of Places, in SIGIR Workshop on Geographic Information Retrieval, pages 14–16, 2006.See http://mmis.doc.ic.ac.uk/www-pub/sigir06-GIR.pdf.
- Simone Paolo Ponzetto and Michael Strube, 2006. Exploiting Semantic Role Labeling, WordNet and Wikipedia for Coreference Resolution, in NAACL 2006. See http://www.eml-research.de/english/homes/strube/papers/naacl06.pdf.
- Simone Paolo Ponzetto and Michael Strube, 2007a. Deriving a Large Scale Taxonomy from Wikipedia, in Association for the Advancement of Artificial Intelligence (AAAI2007).
- Simone Paolo Ponzetto and Michael Strube, 2007b. Knowledge Derived From Wikipedia For Computing Semantic Relatedness, in Journal of Artificial Intelligence Research 30 (2007) 181-212. See also these PPT slides (in PDF format): Part I, Part II, and References.
- Simone Paolo Ponzetto and Michael Strube, 2007c. An API for Measuring the Relatedness of Words in Wikipedia, in Companion Volume to the Proceedings of the 45th Annual Meeting of the Association for Computational Linguistics, See pages 49-52 within http://acl.ldc.upenn.edu/P/P07/P07-2.pdf
- Simone Paolo Ponzetto, 2007. Creating a Knowledge Base from a Collaboratively Generated Encyclopedia, in Proceedings of the NAACL-HLT 2007 Doctoral Consortium, pp 9-12, Rochester, NY, April 2007. See http://www.aclweb.org/anthology-new/N/N07/N07-3003.pdf.
R – S
- Tyler Riddle. 2006. Parse::mediawikidump. URL http://search.cpan.org/~triddle/Parse-MediaWikiDump-0.40/.
- Ralf Schenkel, Fabian Suchanek and Gjergji Kasneci, 2007. YAWN: A Semantically Annotated Wikipedia XML Corpus, in BTW2007.
- Péter Schönhofen, 2006. Identifying Document Topics Using the Wikipedia Category Network, in Proceedings of the 2006 IEEE/WIC/ACM International Conference on Web Intelligence, pp. 456-462, 2006.See http://amber.exp.sis.pitt.edu/gale/paper/Identifying%20document%20topics%20using%20the%20Wikipedia%20category%20network.ppt.
- Börkur Sigurbjörnsson, Jaap Kamps, and Maarten de Rijke. 2006. Focused Access to Wikipedia. In Proceedings DIR-2006.
- Suzette Kruger Stoutenburg, 2007. Research Proposal: Acquiring the Semantic Relationships of Links between Wikipedia Articles, class proposal to http://www.coloradostoutenburg.com/cs589%20stoutenburg%20project%20proposal%20v1.doc
- Strube, M. and Ponzetto, S. P. 2006. WikiRelate! Computing Semantic Relatedness Using Wikipedia, in AAAI, pages 1419-1424, Boston, MA.
- Fabian M. Suchanek, Gjergji Kasneci and Gerhard Weikum, Yago – A Core of Semantic Knowledge, in 16th international World Wide Web Conference (WWW 2007), Banff, Alberta. See http://www.mpi-inf.mpg.de/~suchanek/downloads/yago/.
- Fabian M. Suchanek, Gjergji Kasneci and Gerhard Weikum, 2007. Yago: A Large Ontology from Wikipedia and WordNet, in Technical Report, submitted to the Elsevier Journal of Web Semantics 67 pp. See http://www.mpi-inf.mpg.de/~suchanek/publications/yagotr.pdf.
- Fabian M. Suchanek, Georgiana Ifrim and Gerhard Weikum, 2006. Combining Linguistic and Statistical Analysis to Extract Relations from Web Documents, in Knowledge Discovery and Data Mining (KDD 2006). See http://www.mpi-inf.mpg.de/~ifrim/publications/kdd2006.pdf.
- S. Suh, H. Halpin and E. Klein, 2006. Extracting Common Sense Knowledge from Wikipedia, in Proc. International Semantic Web Conference 2006 Workshop, Web Content Mining with Human Language Technologies, Athens, GA, 2006. See http://orestes.ii.uam.es/workshop/22.pdf.
- Zareen Syed, Tim Finin, and Anupam Joshi, 2008. Wikipedia as an Ontology for Describing Documents, from Proceedings of the Second International Conference on Weblogs and Social Media, AAAI, March 31, 2008. See http://ebiquity.umbc.edu/paper/html/id/383/Wikipedia-as-an-Ontology-for-Describing-Documents.
T – V
- J. A. Thom, J, Pehcevski and A. M. Vercoustre, 2007. Use of Wikipedia Categories in Entity Ranking. in Proceedings of the 12th Australasian Document Computing Symposium, Melbourne, Australia (2007). See http://arxiv.org/PS_cache/arxiv/pdf/0711/0711.2917v1.pdf.
- Antonio Toral and Rafael Muñoz, 2007. Towards a Named Entity Wordnet (NEWN), in Proceedings of the 6th International Conference on Recent Advances in Natural Language Processing (RANLP). Borovets (Bulgaria). pp. 604-608 . September 2007.See http://www.dlsi.ua.es/~atoral/publications/2007_ranlp_newn_poster.pdf.
- Antonio Toral and Rafael Muñoz, 2006. A Proposal to Automatically Build and Maintain Gazetteers for Named Entity Recognition by using Wikipedia, in Workshop on New Text, 11th Conference of the European Chapter of the Association for Computational Linguistics, Trento (Italy). April 2006.See http://www.dlsi.ua.es/~atoral/publications/2006_eacl-newtext_wiki-ner_paper.pdf.
- Anne-Marie Vercoustre, Jovan Pehcevski and James A. Thom, 2007. Using Wikipedia Categories and Links in Entity Ranking, in Pre-proceedings of the Sixth International Workshop of the Initiative for the Evaluation of XML Retrieval (INEX 2007), Dec 17, 2007. See http://hal.inria.fr/docs/00/19/24/89/PDF/inex07.pdf.
- Anne-Marie Vercoustre, James A. Thom and Jovan Pehcevski, 2008. Entity Ranking in Wikipedia, in SAC’08 March 16-20, 2008, Fortaleza, Ceara, Brazil. See http://arxiv.org/PS_cache/arxiv/pdf/0711/0711.3128v1.pdf.
- Max Völkel, Markus Krötzsch, Denny Vrandecic, Heiko Haller and Rudi Studer, 2006. Semantic Wikipedia, in Proceedings of WWW2006, pp 585-594.See http://www.aifb.uni-karlsruhe.de/WBS/hha/papers/SemanticWikipedia.pdf
- Jakob Voss. 2006. Collaborative Thesaurus Tagging the Wikipedia Way. ArXiv Computer Science e-prints, cs/0604036. See http://arxiv.org/abs/cs.IR/0604036
- Denny Vrandecic, Markus Krötzsch and Max Völkel, 2007. Wikipedia and the Semantic Web, Part II, in Phoebe Ayers and Nicholas Boalch, Proceedings of Wikimania 2006 – The Second International Wikimedia Conference, Wikimedia Foundation, Cambridge, MA, USA, August 2007.See http://wikimania2006.wikimedia.org/wiki/Proceedings:DV1.
W – Z
- Wang Y , Wang H , Zhu H , Yu Y, 2007. Exploit Semantic Information for Category Annotation Recommendation in Wikipedia, in Natural Language Processing and Information Systems (2007), pp. 48-60.
- Gang Wang, Yong Yu and Haiping Zhu, 2007. PORE: Positive-Only Relation Extraction from Wikipedia Text, at ISWC 2007, See http://iswc2007.semanticweb.org/papers/ISWC2007_RT_Wang(1).pdf.
- Yotaro Watanabe, Masayuki Asahara and Yuji Matsumoto, 2007. A Graph-based Approach to Named Entity Categorization in Wikipedia Using Conditional Random Fields, in EMNLP-CoNLL 2007, 29th June 2007, Prague, Czech. Has a useful CRF overview. See http://www.aclweb.org/anthology-new/D/D07/D07-1068.pdf and http://cl.naist.jp/~yotaro-w/papers/2007/emnlp2007.ppt.
- Timothy Weale, 2006. Utilizing Wikipedia Categories for Document Classification.
- Nicolas Weber and Paul Buitelaar, 2006. Web-based Ontology Learning with ISOLDE. See http://orestes.ii.uam.es/workshop/4.pdf.
- Wikify!, online service to automatically turn selected keywords into Wikipedia-style links. See http://www.wikifyer.com/.
- Wikimedia Foundation. 2006. Wikipedia. URL http://en.wikipedia.org/wiki/Wikipedia:Searching.
- Wikipedia, full database. See http://download.wikimedia.org.
- Wikipedia, general references. See especially (but only a small subset) for the Semantic Web, RDF, RDF Schema, OWL, SPARQL, GRDDL, W3C, Linked Data, many, many ontologies and controlled vocabularies such as FOAF, SKOS, SIOC, Dublin Core, etc; description logic areas (such as FOL), etc., etc. etc.
- Wikipedia, (as a) research source; see http://en.wikipedia.org/wiki/Wikipedia:Wikipedia_in_academic_studies.
- Fei Wu and Daniel S. Weld, 2007. Autonomously Semantifying Wikipedia.
- Hugo Zaragoza, Henning Rode, Peter Mika, Jordi Atserias, Massimiliano Ciaramita & Giuseppe Attardi, 2007. Ranking Very Many Typed Entities on Wikipedia, in CIKM ’07: Proceedings of the Sixteenth ACM International Conference on Information and Knowledge Management. See http://grupoweb.upf.es/hugoz/pdf/zaragoza_CIKM07.pdf.
- Torsten Zesch, Iryna Gurevych, Max Mühlhäuser, 2007. Analyzing and Accessing Wikipedia as a Lexical Semantic Resource, and the longer technical report. See http://www.ukp.tu-darmstadt.de/software/JWPL.
- Torsten Zesch and Iryna Gurevych, 2007. Analysis of the Wikipedia Category Graph for NLP Applications, in Proceedings of the TextGraphs-2 Workshop (NAACL-HLT).
- Vinko Zlatic, Miran Bozicevic, Hrvoje Stefancic, and Mladen Domazet. 2006. Wikipedias: Collaborative Web-based Encyclopedias as Complex Networks, in Physical Review E, 74:016115.





 I still never cease to be amazed at how wonderful and powerful tools are so often and easily overlooked. The most recent example is
I still never cease to be amazed at how wonderful and powerful tools are so often and easily overlooked. The most recent example is